1.Introduction
Protein degradation at the spatial and temporal regulation is essential for many cellular processes, including cell cycle progression, signaling, differentiation, and growth, whereas its dysregulation has been implicated in almost all hallmarks of cancer. The rapid development of technologies for targeted protein degradation, such as proteolytic targeted chimeras (PROTACs), enabled many previously non-druggable proteins, providing new insights for drug discovery and design1. Over 80% of intracellular protein degradation in cells is mainly regulated by the ubiquitin-proteasome system (UPS). E3 ubiquitin ligases and degrons, which are short linear motifs embedded within the sequences of modular proteins used by E3 ligases to target proteins, represent the fundamental parts of the UPS. A key property of degron is transferability, i.e., in most cases, transplanting degron from an unstable protein onto another protein accelerates the degradation of that protein, which makes it promising for targeted protein degradation. In this work, we first systematically analyzed the multimodal biometrics of degron. We found sequences around degron are more evolutionarily conserved, and significantly more phosphorylation and ubiquitination sites are enriched. Structurally, degron was more likely to be located in protein structure regions with high disorder, high solvent accessibility, poor stability and weak rigidity. Based on these findings, wHere, we build a user-friendly web service, named MetaDegron (Multiple feature integrated Transformer) for E3 degron binding prediction. The built-in MetaDegron model shows excellent performance with AUC value >0.89 by integrating comprehensive featurization strategies and large protein language models. MetaDegron will serve the community for exploring biological mechanisms and implications of protein degradation, as well as drug discovery and design on degrons.
2. Model
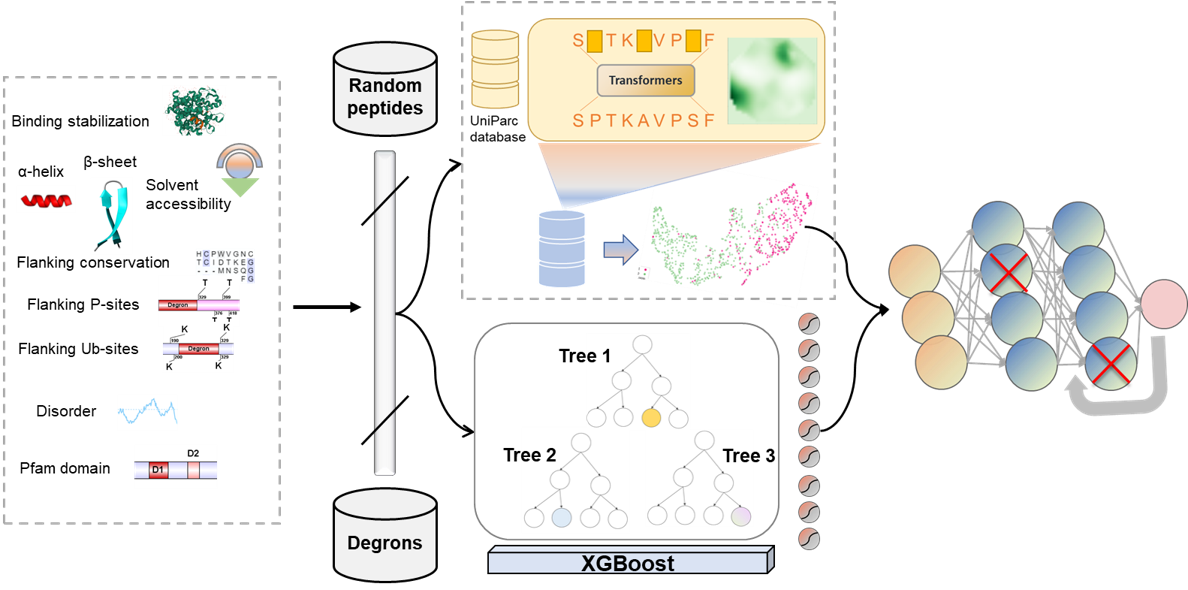
Figure 1. The workflow of MetaDegron for E3 targeted degron prediction.
First, multiple characteristic features, i.e., disorder, solvent accessibility and secondary structure (COIL, HELIX and SHEET), rigidity, stabilization upon binding, flanking conservation, structured domains, degron-associated phosphorylation sites, degron-associated ubiquitinated lysines, were calculated for each degron instances and random peptides. All characteristic features were evaluated by a XGBoost classifier trained on known degron instances. Moreover, for embedding of degrons and their surrounding sequence, we employed a pre-trained transformer model (SeqVec) to represent each degron by 1024-dimensional embedding. SeqVec was implemented based on the language model using the deep bi-directional LSTM (BLSTM) architecture for protein sequences transferring the knowledge obtained by predicting the next amino acid in 33 million proteins (UniRef50). These representations are capable of accurately depicting the biochemical features for each amino acid. Finally, we leveraged the trained numeric vector encodings of degron instances and all characteristic features for learning the bingding between E3 and degron. We constructed a fully connected deep-learning network based on the output of these two submodels, leading to a final layer with a single neuron for predicting the E3 targeted degron.
3. Model performance
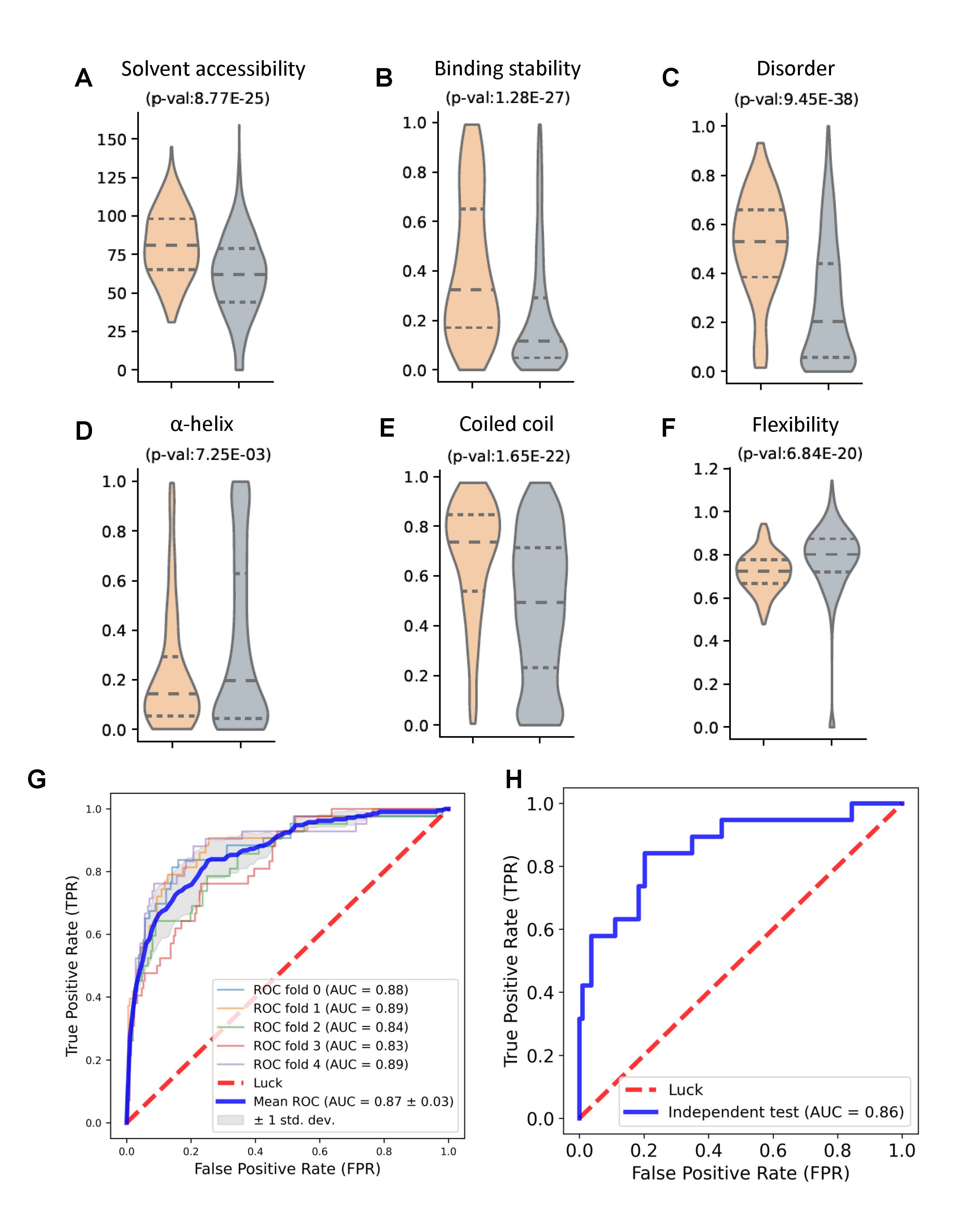
Figure 2. Implementation and validation of MetaDegron performance.
Remarkably, the known degrons exhibited a higher degree of solvent accessibility and binding stability compared to the random peptides (Figure 2A, 2B), suggesting their importance in recognition by degradative enzymes. Furthermore, they were found to be preferentially located in protein disordered regions (Figure 2C), highlighting their distinctive localization patterns. Additionally, the analysis revealed a specific preference of degrons for coiled coil regions rather than α-helix regions (Figure 2D, 2E). It was also observed that degrons tend to occur in lower flexibility regions (Figure 2F). These findings provided valuable insights into the structural characteristics of degrons and indicate potential determinants for degron recognition and degradation. Subsequently, the XGBoost classifier (called MetaDegron-X) was constructed using these discerning features for E3 targeted degron. The performance of MetaDegron-X, as assessed by the AUC values, was promising. Specifically, the AUC values ranged from 0.81 to 0.90 in a five-fold cross-validation, with an average AUC value of 0.87 (Figure 2G). Furthermore, validation of the developed MetaDegron-X was carried out on an independent testing dataset. The performance of MetaDegron-X was superior, as denoted by the AUC value of 0.86 (Figure 2H).
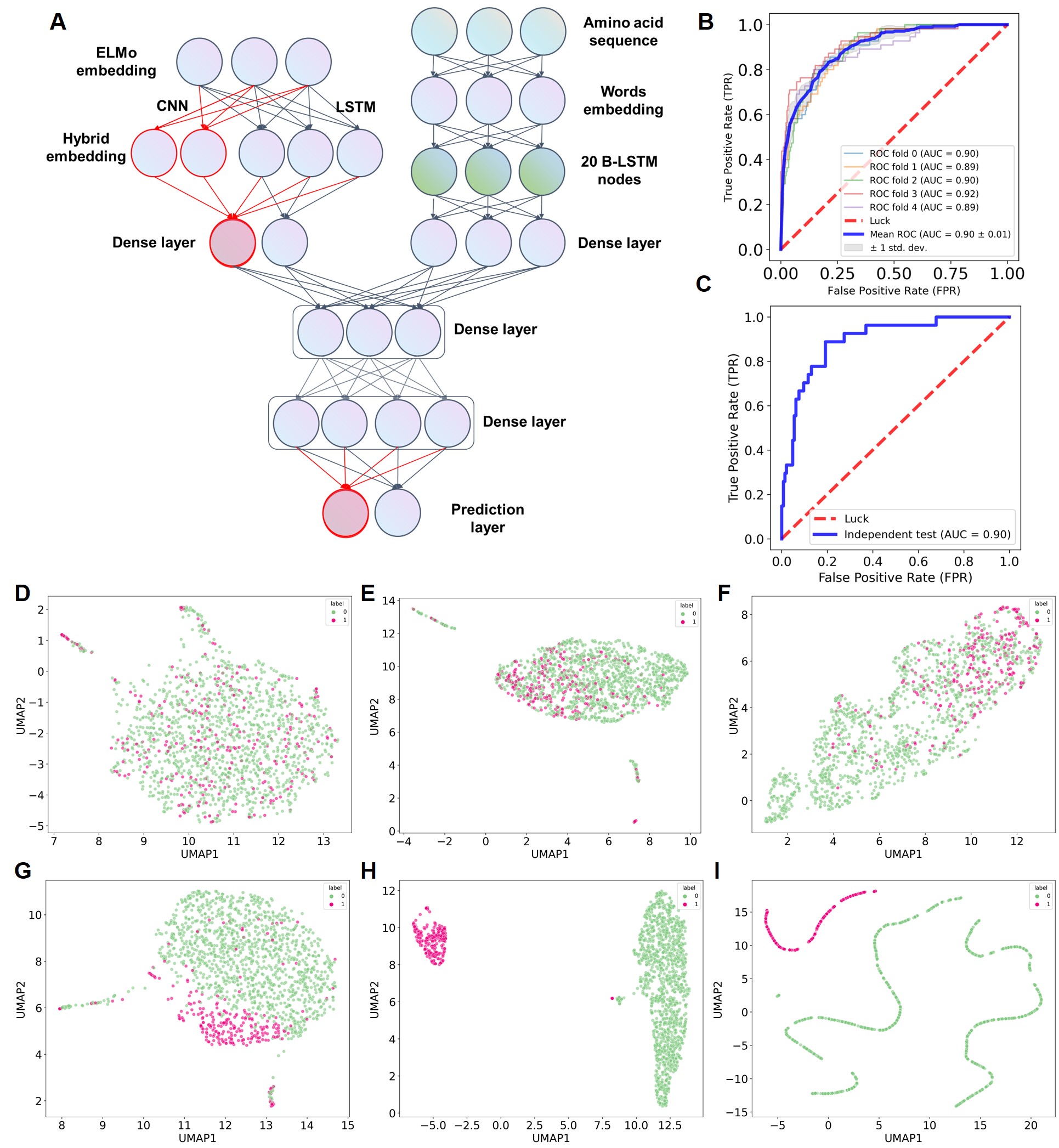
Figure 3. Implementation and validation of MetaDegron performance.
By incorporating a deep learning framework, MetaDegron-D was capable of solely operating on protein sequences. This novel approach utilized a hybrid architecture comprising cutting-edge deep learning networks (Figure 3A), such as protein language models, word embeddings, convolution, and BLSTM, as thoroughly detailed in the methodology section. This deep learning framework allowes MetaDegron-D to leverage the full potential of these advanced networks and their ability to extract high-level features from protein sequences. The performance evaluation of MetaDegron-D demonstrated its great predictive capabilities. Through a five-fold CV approach, we obtained an average AUC value of 0.90. Furthermore, the AUC values ranged from 0.89 to 0.92, indicating consistent and reliable performance (Figure 3B). Additionally, when tested with an independent dataset, MetaDegron-D achieved an improved AUC value of 0.90 (Figure 3C).
To further explore the capabilities of the MetaDegron framework, we employed the method implemented in Becht et al. (Becht et al., 2019) to visualize the degrons and random peptides based on their features at each network layer (Figure 3D-I). As expected, the feature representations of the input layer for both degrons and random peptides exhibited significant overlap and mixing (Figure 3D). However, as the framework underwent training, a clear distinction between degrons and random peptides emerged, resulting in more separated clusters within the feature space (Figure 3H, 3I).
4. General webserver pipeline:
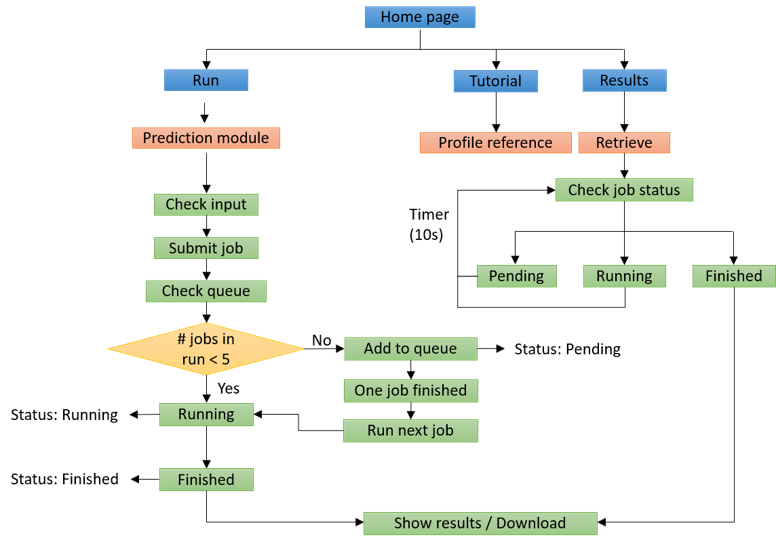
Figure 4. General pipeline for MetaDegron server.
Our webserver provides user-friendly interfaces for users to submit jobs, check job status, and retrieve results.
5. Usage:
Input
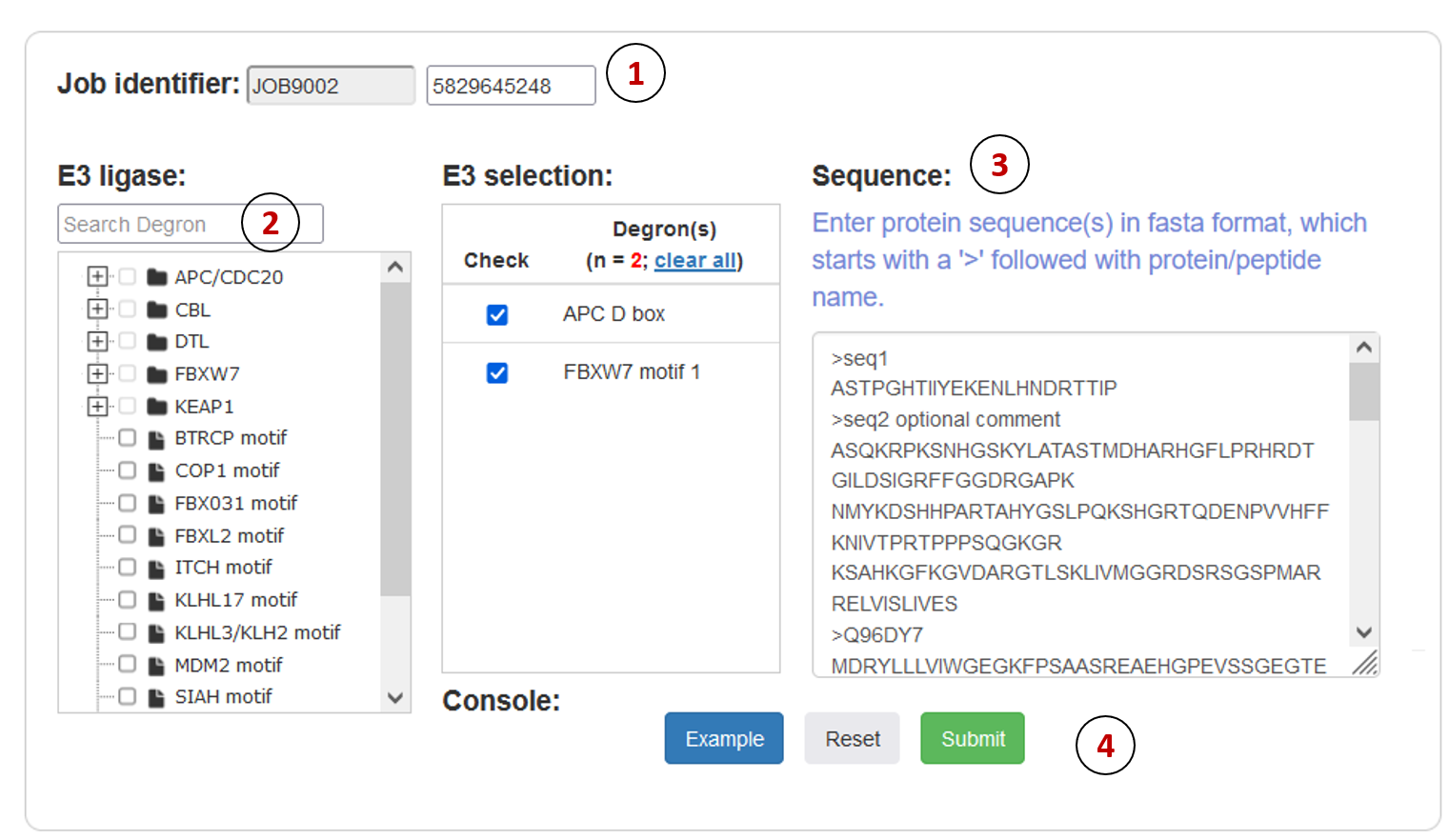
Figure 5. Job submission form.
- Job identifier: Job identifier can be generated automatically or customized by the submitter. It is confidential to other users and can be used for job status monitoring and result retrieval.(See Results page).It is required.
- E3 ligase: The MetaDegron 1.0 server supports 21 E3 ligase prediction. We constructed a classification tree of E3 ligase. Users can quickly retrieve and submit candidate E3 through the search box and tree map.
- Sequence: User can directly copy one or more proteins with FASTA format in the input box.
- Operation buttons: Submit, reset the submission form, or access the example dataset.
Output
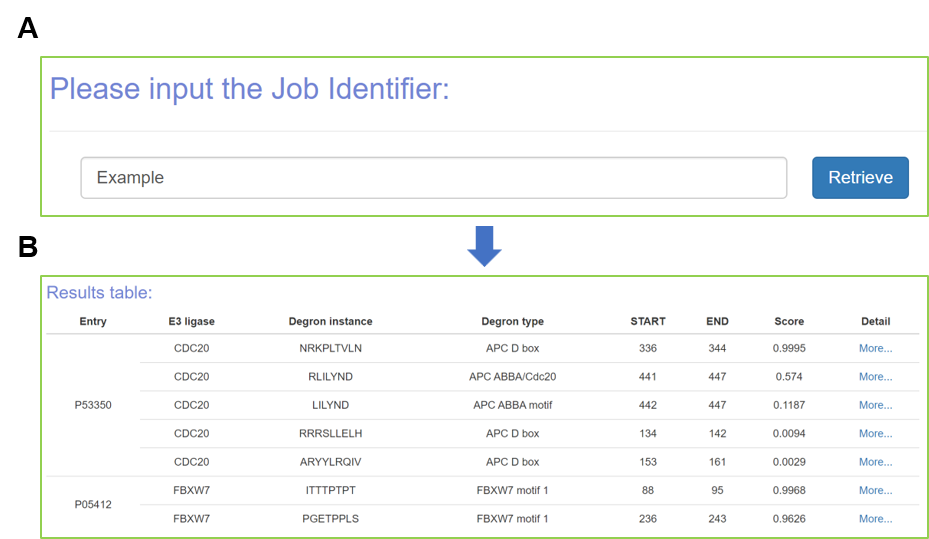
Figure 6. The prediction output.
After finishing the submitted job, the prediction results will be visualized with specific information, including the “Entry”, “E3 ligase”, “Degron instance”, “Degron type”, “Start”, “End”, and “Score” (Figure 6B). It displays the detailed information for degron and source protein (Figure 7). The properties of degron (Figure 7A) and information of source protein (Figure 7B) are displayed as well. In addition, the structure of source protein is presented with 3Dmol.js (Rego and Koes, 2015), and the degron instance is marked with highlights. Moreover, the multiple sequence alignment (MSA) of degron instance and source protein are visualized by using the ProViz tool (Jehl et al., 2016) (Figure 7C), and the interacting E3s or deubiquitinating enzymes (DUBs) of source protein are provided in a tabular list and an interactive network based on the Cytoscape.js (Franz et al., 2023) (Figure 7D).
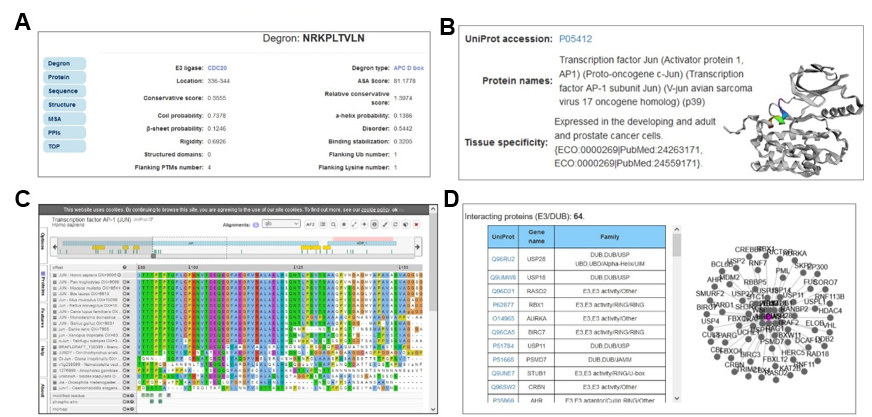
Figure 7. The feature properties of selected degron and the annotations of source protein.
6. Citation:
Please cite:
Zheng M, Lin S, Chen K, Hu R, Wang L, Zhao Z, Xu H. MetaDegron: multimodal feature-integrated protein language model for predicting E3 ligase targeted degrons. Brief Bioinform. 2024 Sep 23;25(6):bbae519. [PMID: 39431517]
Xu H, Hu R, Zhao Z. DegronMD: Leveraging Evolutionary and Structural Features for Deciphering Protein-Targeted Degradation, Mutations, and Drug Response to Degrons. Mol Biol Evol. 2023;40(12):msad253. [PMID: 37992195]
